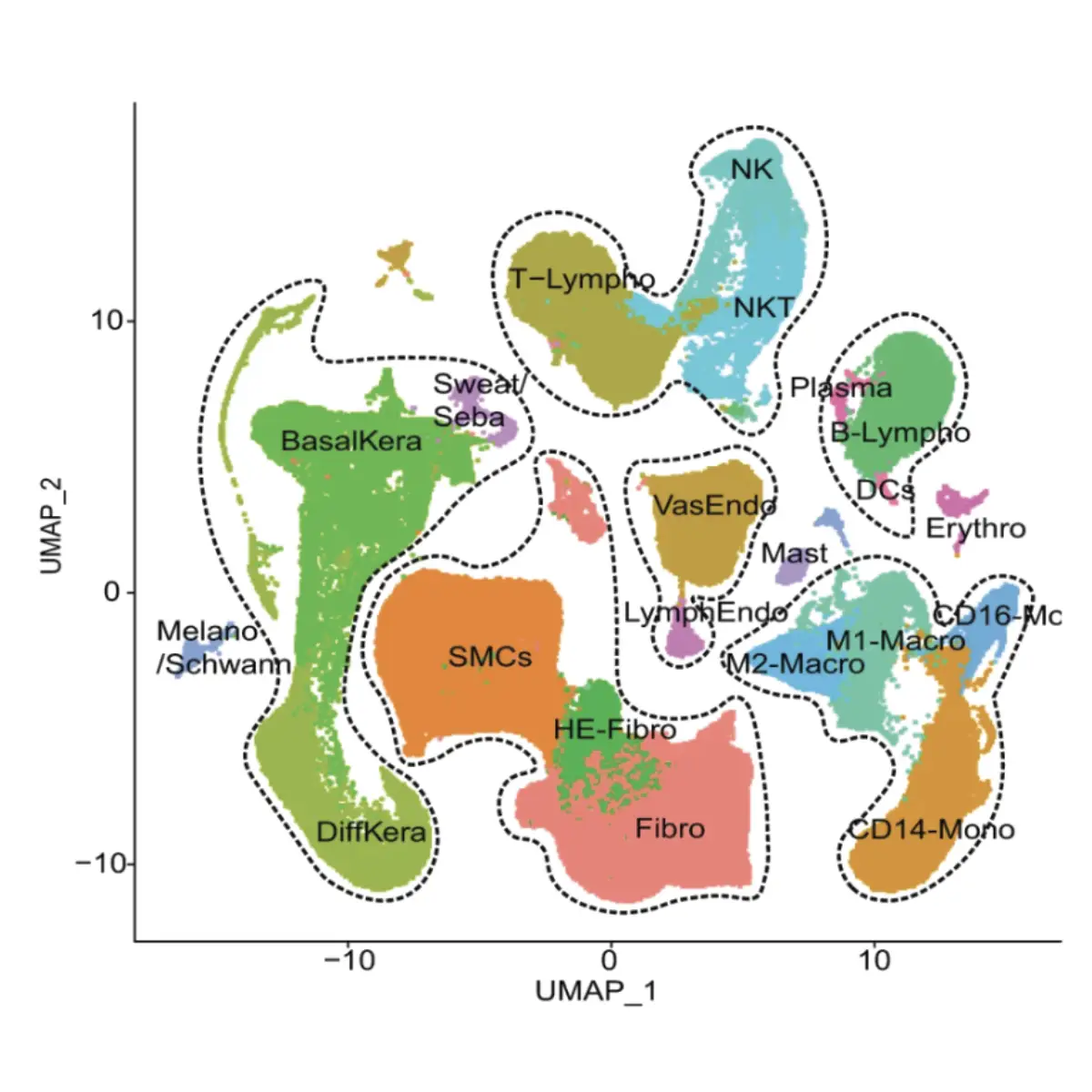
Diabetic foot ulcers (DFUs) are a critical concern because they often lead to severe infections and, in extreme cases, may necessitate amputation. Thus, timely diagnosis, appropriate wound care, and proper management are crucial. By single-cell RNA sequencing analysis, our research delves into the wound microenvironment and molecular mechanisms of the healing process of DFU. Notably, we have identified a unique population of fibroblasts, termed HE-Fibro, highly expressing healing enrichment genes associated with immune and inflammatory response. These findings provide valuable insights into potential avenues for enhancing DFU healing by targeting their unique genetic signatures.
Lab Members Involved:
Relevant Publications
Nature Communications, 2023
Diabetic foot ulceration (DFU) is a devastating complication of diabetes whose pathogenesis remains incompletely understood. Here, we profile 174,962 single cells from the foot, forearm, and peripheral blood mononuclear cells using single-cell RNA sequencing. Our analysis shows enrichment of a unique population of fibroblasts overexpressing MMP1, MMP3, MMP11, HIF1A, CHI3L1, and TNFAIP6 and increased M1 macrophage polarization in the DFU patients with healing wounds. Further, analysis of spatially separated samples from the same patient and spatial transcriptomics reveal preferential localization of these healing associated fibroblasts toward the wound bed as compared to the wound edge or unwounded skin. Spatial transcriptomics also validates our findings of higher abundance of M1 macrophages in healers and M2 macrophages in non-healers. Our analysis provides deep insights into the wound healing microenvironment, identifying cell types that could be critical in promoting DFU healing, and may inform novel therapeutic approaches for DFU treatment. More information: https://doi.org/10.1038/s41467-021-27801-8





Relevant Presentations
Atlanta Workshop for Single-cell Omics, 2023
Diabetic foot ulcers (DUs) develop in over 15% of diabetic patients during their lifetime and are the initial event in 85% of major amputations that are performed in people with diabetes. We recently identified a unique population of fibroblasts with over-expression of inflammatory and matrix remodeling genes that are lacking in DFUs with impaired healing. To further explore the regulatory space of DUs and identify the long non-coding RNAs (IncRNAs) associated with the healing of DUs, we conducted comprehensive single-cell data analysis. The IncRNAs are key regulators of the genes and are currently overlooked in the traditional analysis due to the lack of a well-annotated non-coding RNA genome. To address this issue, we have developed a newIncRNAs analysis workflow by collecting the annotation information from databases including IncRNAKB and LncExpDB. The combination of IncRNAs information from these databases yielded a comprehensive annotation file with over 80,000 entries that are currently missing in the ENCODE annotation. The single-cell data from 54 samples from foot, forearm, and peripheral blood mononuclear cells was analyzed using cell ranger with this enhanced annotation to generate IncRNAs expression profiles. Samples belong to four clinical groups including non- diabetic, diabetic, DFU healer, and DFU non-healer. In addition to IncRNA-focused analysis, we also performed an integrated analysis of IncRNA and genes to identify the gene-regulatory modules that are significantly associated with DFU healing. Our exploration of the IncRNAs in the DFU samples revealed that the expression levels of IncRNAs are significantly lower than those of genes. Normalization and clustering of single-cell data based on IncRNAs generated well-segregated clusters corresponding to different cell types like our previously published gene expression-based data. Notably, the IncRNA-based clustering provided better resolution for major cell types including SMCs and Healer Enriched Fibroblasts while lowering the resolution for small clusters such as plasmas and erythrocytes. Further analysis depicted that HE-Fibro is significantly enriched in DFU healers (P<0.05) with over-expression of multiple immune, angiogenesis, and matrix remodeling IncRNAs including HSALNG0057306, HSALNG0109104, HSALNG0070210, HIF1A-AS1, HSALNG0137013, HSALNG0132877, and HSALNG0106482. Further de-novo network analysis based on the expression of IncRNA-gene identified multiple healer-enriched modules related to cell junctions, complement activation, collagen fibril organization, and neuroinflammation. Based on our knowledge this is the first study analyzing the gene-regulatory landscape of D U s by studying IncRNAs at the single-cell level to understand the mechanism of ulcer healing. The study identified multiple novel IncRNAs as potential therapeutic targets for the treatment of impaired ulcer healing.
- Venue: Atlanta Workshop for Single-cell Omics 2023



Atlanta Workshop for Single-cell Omics, 2023
Diabetic foot ulcer (DU) is a fatal complication of diabetes that affect 15% of diabetic patients during their lifetime. DFU significantly degrades patients' quality of life and can lead to lower limb amputation and even mortality. Given that the diabetes population has been increasing, it is important to unveil the pathogenesis and healing process of DU. However, little has been known about how various types of cells such as fibroblast and immune cells are involved in the process of wound healing. Methods: We profiled 28,140 cells by single-cell RNA-Sequencing from 7 patient samples (4 DFU-Healer and 3 DFU-Non-healer) after optimizing the single-cell profiling of wound debridement that generally yields poor quality cells. The gene expression matrix was used to perform dimensionality reduction by PCA and UMAP and to cluster cells by a graph-based clustering approach. Intercellular communication networks and gene regulatory networks (GRN) were inferred and analyzed to find genes and transcription factors that correlate with the healing outcome of the ulcers. Then pathway enrichment analysis was performed on the genes associated with healing to understand underlying molecular mechanisms. Results: Single-cell profiling on the wound debridement samples captured the profile of 28,140 cells from 15 different cell types based on the expression of established marker genes. The comparative analysis of cellular proportion depicted significantly higher enrichment of healing-enriched fibroblasts in the DFU healers as compared to non-healers (P=0.05). These healing-enriched fibroblasts showed gene expression profiles that are associated with immune/inflammatory response (CHI3L1, HIF-1 a, IL-1) and remodeling the extracellular matrix (MMP1, ASP). Further gene-regulatory module analysis on healing-enriched fibroblasts depicted significant activation of NFKB1, SOX4, and FOX1 transcriptional factors and target genes indicating the role of these cells in mounting inflammatory response. Additionally, comparative analysis of macrophages revealed a significantly higher abundance of M2-macrophage in the DFU-Healers as compared to non-healer. These M2 macrophages in the healers also depicted activation of MAFB, SPI1 and TFEC transcription factors. Further cellular communication analysis depicted that healing-enriched fibroblasts are key communicators of molecular signals among the cell types. Conclusions: The study addressed a critical need by successfully mapping the single-cell landscape of DFU from the routine debridement samples as compared to surgical samples reported in previous studies. The study identified higher enrichment of healing-enriched fibroblasts in the wounds that healed in 12 weeks indicating their potential role in healing. In the future interventions that help in the enrichment of these healing-enriched fibroblasts might promote faster healing to reduce the complications due to impaired wound healing.
- Venue: Atlanta Workshop for Single-cell Omics 2023



Atlanta Workshop for Single-cell Omics, 2023
Diabetic foot ulcers (DFUs) are a severe complication among diabetic patients and often result in amputation, and even mortality. Early recognition of infection and ischemia is crucial for improved healing, but current methods are invasive, time-consuming, and expensive. To address this need, we have developed DFUCare, a workflow that uses computer vision and deep learning algorithms to non- invasively localize, classify, and analyze DFUs. The workflow uses a combination of an L*a*b and Y*Cr*Cb color space segmentation with a pre-trained YOLOv5s algorithm for wound localization achieving an F1 score of 0.80 and an mAP of 0.861 on a test set. Using deep-learning algorithms to identify infection and ischemia, we achieved a binary accuracy of 89.01 % for infection classification and 88.74% for ischemic classification on a validation set. DFUCare also quantifies wound size, performs tissue color and textural analysis to allow comparative analysis of macroscopic features of wound. We tested the workflow performance in a clinical setting to analyze the DFUs collected using a cell phone on which workflows successfully segmented from the background from the skin, localized the wound and predicted infection and ischemia. This innovative approach has the potential to deliver a paradigm shift in diabetic foot care by providing a cost-effective, remote, and convenient healthcare solution.











