In contrast to single-cell transcriptomics, spatial transcriptomics approaches preserve the localized gene expression across tissue architectures, illuminating cellular and genomic heterogeneity in a histological context. However, cellular resolution remains a significant limitation of unbiased whole transcriptome spatial methodologies, wherein each capture spot contains the transcripts of 1-10 cells rather than providing single-cell resolution. To overcome this limitation, great attention has been drawn to integrated single-cell and spatial transcriptomics methodologies, in which the spatial transcriptomic profiles of tissues can be deconvoluted using single-cell transcriptomic data. The ongoing work in the Bhasin Lab leverages this integrated approach to elucidate the cellular and transcriptomic landscapes of the tissues over the pathogenesis of vascular disease and cancers.
Lab Members Involved:
Relevant Publications
An Integrated Single-Nuclei and Spatial Transcriptomic Analysis for Investigating Acute Distension Injury in Vein Graft Implantation
Circulation Research, 2023
Circulation Research, 2023
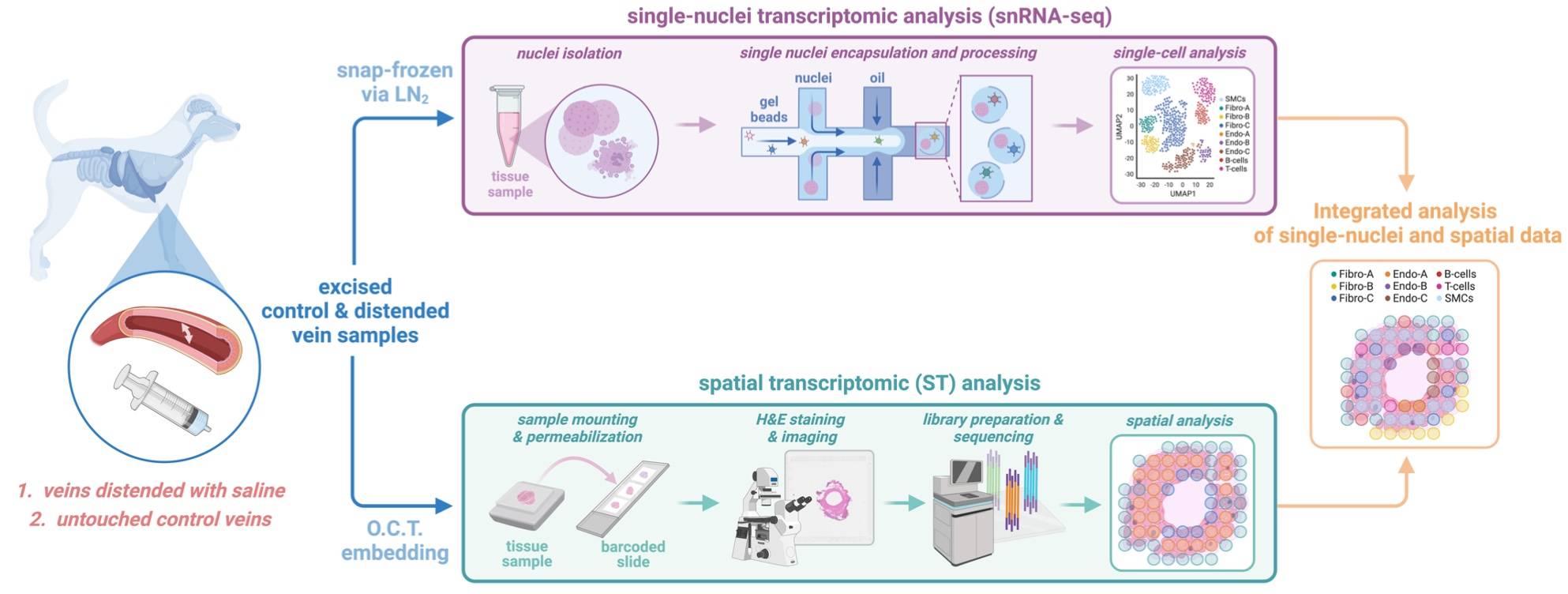
Autogenous vein is the most commonly used conduit for coronary and lower extremity arterial bypass grafting. Standard vein harvest technique involves pressure distention and manipulation of the vein conduit, and this process has been suspected to elicit early cell injury and lead to an increased risk of bypass graft failure. Utilizing single-nuclei and spatial transcriptomics, we profiled the genomic effects of distension during graft harvesting in a canine model. Saline distention of the vein graft resulted in acute injury and dysregulation of genes associated with extracellular matrix remodeling, cell proliferation and migration, and endothelial-mesenchymal transition (End MT). We further identified distinct subpopulations of endothelial cells (ECs) and fibroblasts (FBs) with disparate transcriptomic programs related to endothelial injury, EC activation, FB activation, and EndMT, which were present in altered proportions in distended veins compared to control veins. Lastly, we examined the spatial distribution and intercellular communication networks between the various cellular populations to elucidate the cellular and transcriptomic landscape of the vein following distension. The work presented herein represents the first applications of single-nuclei and spatial transcriptomic analyses to the investigation of vein graft implantation. Through this approach, we have unveiled a prompt genomic response to distension amongst distinct cellular populations, which were non-uniformly distributed throughout specific layers of vein, thus providing new molecular insights critical for guiding strategies to limit vein harvest injury.






Relevant Presentations
Investigation of Acute Distension Injury in vein Graft Implanation Through Integrated Single-Nuclei and Spatial Transcriptomic Analysis
Vascular Research Initiatives Conference, 2023
Vascular Research Initiatives Conference, 2023
Autogenous vein is the most commonly used conduit for coronary and lower extremity arterial bypass grafting. Standard vein harvest technique involves pressure distention and manipulation of the vein conduit, and this process has been suspected to elicit early cell injury and lead to an increased risk of bypass graft failure. Utilizing single-nuclei and spatial transcriptomics, we profiled the genomic effects of distension during graft harvesting in a canine model. Saline distention of the vein graft resulted in acute injury and dysregulation of genes associated with extracellular matrix remodeling, cell proliferation and migration, and endothelial-mesenchymal transition (End MT). We further identified distinct subpopulations of endothelial cells (ECs) and fibroblasts (FBs) with disparate transcriptomic programs related to endothelial injury, EC activation, FB activation, and EndMT, which were present in altered proportions in distended veins compared to control veins. Lastly, we examined the spatial distribution and intercellular communication networks between the various cellular populations to elucidate the cellular and transcriptomic landscape of the vein following distension. The work presented herein represents the first applications of single-nuclei and spatial transcriptomic analyses to the investigation of vein graft implantation. Through this approach, we have unveiled a prompt genomic response to distension amongst distinct cellular populations, which were non-uniformly distributed throughout specific layers of vein, thus providing new molecular insights critical for guiding strategies to limit vein harvest injury.












